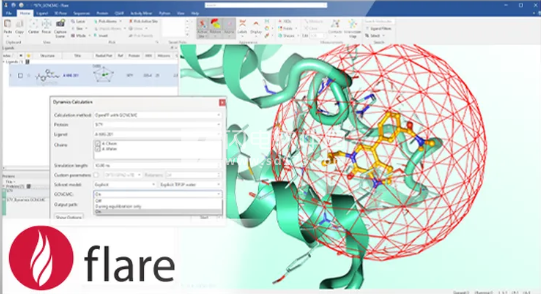
Cresset-BMD Flare是用于基于配体和结构的药物设计的绝佳解决方案,让药物研究、开发设计人士能够仔细地检查配体,深入了解其蛋白质靶标和配体,放心地进行先导化合物优化。现在用户不需要花费大量的时间就能获得优化的结果,更具信心地制定决策,准确地计算和简单地操作,便捷地沟通,Flare V10在最新解决方案中带来Spark生物等排体替换、同源建模、集成 MM/GBSA 等一系列新增和改进,为所有用户带来了新的和增强的科学功能和方法。
Capabilities in Flare V10 include:
Smart import and handling of protein-ligand complexes
Accurate and reliable protein preparation
Ligand preparation, including enumeration of stereo centers, stripping of salts, protonation at pH 7 and tautomer enumeration
Sequence alignment and superimposition
Accurate docking using Lead Finder
Advanced ligand-based conformation hunt and alignment
Pharmacophore building with FieldTemplater
Electrostatic Complementarity maps and score
Protein interaction potentials
Qualitative Structure-Activity Relationship analysis using Activity Atlas and Activity Miner
Quantitative SAR models of regression and classification using Field QSAR and machine learning methods
Enumeration to generate libraries and arrays based on more than 100 popular synthetic chemistry reactions
Chemical exploration of a hit or lead compound using Hit Expander
R-Group Decomposition and Analysis
Scaffold hopping and bioisostere replacement using Spark
Molecular Mechanics/Generalized Born Surface Area (MM/GBSA) calculations of ligand-protein binding free energy on individual protein conformations or on conformations ensembles from Molecular Dynamics
Quantum Mechanics calculations on ligands
Free Energy Perturbation simulations
Support for the Open Force Field
Molecular Dynamics
Automatic creation of custom torsional parameters for ligands in support of Dynamics and FEP calculations
3D-RISM water analysis
MD-based analysis of water thermodynamics with GIST
Pocket detection to identify potential drug binding sites in the protein targets of interest
Homology modeling to create reliable 3D structures for the targets of interest
WaterSwap analysis for ligand and binding site energetics
Minimization of protein ligand complexes using XED and OpenMM
The Flare Python API
基于配体的药物设计者利用 Flare 根据分子的形状、静电和结合活性对其分子进行仔细检查、比较和确定优先级。通过预测新化合物的活性和 ADMET 特性的强大 QSAR 模型,用户可以在整个潜在客户组合中建立信心和理解。
借助多种方法,包括对接和评分、静电互补性™、分子动力学、口袋分析和水分析(GIST 和 3D-RISM),基于结构的药物设计人员可以获得对蛋白质-配体结合的新见解。基于围绕配体和蛋白质静电的既定专有方法,结合我们自己的内部和开源研究,用户能够充分了解其目标结构的特征和相互作用。
借助 Python API,Flare 功能是完全可自定义和可扩展的。无论是通过开发自己的扩展,还是希望包含 Cresset 的预制扩展之一,Flare 的 Python 集成都提供了对各种新功能和专用菜单的访问。Flare 还与 DMTA 协作工具 Torx 无缝连接,以简化整个 DMTA 周期的协作。
 Cresset-BMD Flare v10.0.1
Cresset-BMD Flare v10.0.1
 Cresset-BMD Flare v10.0.1
Cresset-BMD Flare v10.0.1